Difference between revisions of "SEED Viewer Manual/Menu"
| (31 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
== Menu Overview == | == Menu Overview == | ||
| + | |||
| + | The menu is a small bar below the logo of each page. Most menus have submenus that can be accessed via hovering over the menu. Click the submenu entry you want to go to. If no submenu is present, you can directly click the menu itself. | ||
=== Navigate Menu === | === Navigate Menu === | ||
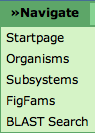
| − | This menu is present on every | + | This menu is present on every SeedViewer page. The first entry '''Startpage''' will lead to the SeedViewer [[SEED_Viewer_Manual|HomePage]]. |
| − | '''Organisms''' will open the [[SEED_Viewer_Manual/ | + | '''Organisms''' will open the [[SEED_Viewer_Manual/OrganismSelect|Organism Select]] page. You will see an overview of all organisms in the SEED and are able to select one to view in detail. |
'''Subsystems''' will do the same for subsystems. | '''Subsystems''' will do the same for subsystems. | ||
| + | |||
| + | '''Scenarios''' lets you browse [[Glossary#Scenarios|Scenarios]] on the [[SEED_Viewer_Manual/Scenarios|Scenarios Page]]. | ||
'''FIGFams''' leads to an entry point for browsing [[SEED_Viewer_Manual/FIGfams|FIGfams]]. You can find more information about FIGfams [[Glossary#FIGfam|here]]. | '''FIGFams''' leads to an entry point for browsing [[SEED_Viewer_Manual/FIGfams|FIGfams]]. You can find more information about FIGfams [[Glossary#FIGfam|here]]. | ||
| Line 14: | Line 18: | ||
[[Image:MenuNav.png]] | [[Image:MenuNav.png]] | ||
| + | |||
| + | === Help Menu === | ||
| + | |||
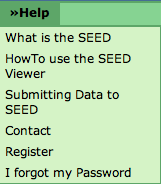
| + | This menu is also present on all SeedViewer pages. The first link '''What is the SEED''' will lead you to the [[Home_of_the_SEED|SEED Homepage]]. | ||
| + | |||
| + | Clicking the second link '''How to use the SEED Viewer''' will get you to this wiki. | ||
| + | |||
| + | '''Submitting data to SEED''' lets you browse the [[RAST_Tutorial|RAST Tutorial]]. | ||
| + | |||
| + | '''Contact''' enables you to write an email to the SEED team. | ||
| + | |||
| + | Clicking '''Register''' is the first step to get a user account. It will lead to the [[SEED_Viewer_Manual/Register|Register]] page. | ||
| + | |||
| + | '''I forgot my password''' enables you to [[SEED_Viewer_Manual/RequestNewPassword|Request a new password]]. | ||
| + | |||
| + | [[Image:MenuHelp.png]] | ||
| + | |||
| + | === Organism Menu === | ||
| + | |||
| + | In the context of a selected organism, an organism menu will appear. '''General Information''' will lead to the [[SEED_Viewer_Manual/OrganismPage|Organism Page]] of the selected organism. | ||
| + | |||
| + | The '''Feature Table''' will open the [[SEED_Viewer_Manual/GenomeBrowser|Genome Browser]] and show you the features present in that organism. | ||
| + | |||
| + | '''Genome Browser''' leads to the [[SEED_Viewer_Manual/GenomeBrowser|Genome Browser]] for the selected organism. | ||
| + | |||
| + | Clicking '''Scenarios''' shows the [[SEED_Viewer_Manual/Scenarios|Scenarios]] page for your organism. | ||
| + | |||
| + | '''Subsystems''' opens a page that lets you select subsystems. | ||
| + | |||
| + | '''Export''' enables you to [[SEED_Viewer_Manual/DownloadOrganism|download]] the features of your organism. | ||
| + | |||
| + | [[Image:MenuOrganism.png]] | ||
| + | |||
| + | === Comparative Tools === | ||
| + | |||
| + | This menu is also present whenever an organism is viewed. Different kinds of comparisons of your selected organism to other organisms are available here. | ||
| + | |||
| + | '''Function based Comparison''' - The [[SEED_Viewer_Manual/CompareMetabolicReconstruction|Compare Metabolic Reconstruction]] will enable you to see the metabolic reconstruction of your selected organism against that of another one. | ||
| + | |||
| + | The '''Sequence based Comparison''' [[SEED_Viewer_Manual/MultiGenomeCompare|Multi Genome Compare]] shows a table and a graphic comparing a selected set of organisms projected against your chosen one (BLAST-based). | ||
| + | |||
| + | Use the '''KEGG Metabolic Analysis''' [[SEED_Viewer_Manual/KEGG|to project the metabolic capabilities]] of your organism onto KEGG maps. | ||
| + | |||
| + | Blasting against your organism is possible using the '''[[SEED_Viewer_Manual/BLASTOrganism|BLAST Search]]'''. | ||
| + | |||
| + | |||
| + | [[Image:CompTools.png]] | ||
| + | |||
| + | === Feature Menu === | ||
| + | |||
| + | Whenever a feature is defined on a SeedViewer page, you will find the '''Feature''' menu for that feature. | ||
| + | |||
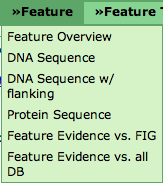
| + | The '''Feature Overview''' points to the [[SEED_Viewer_Manual/Annotation|Annotation]] page. It shows general information about the feature, als well as a Compare Regions View that displays the feature in its genomic context and in comparison to homologs in other genomes. | ||
| + | |||
| + | '''DNA Sequence''' will open a page with the DNA Sequence of the feature (in FASTA format). '''DNA w/ flanking''' not only prints the DNA sequence of the feature, but also includes a user-defined number of bases upstream and downstream of the feature. '''Protein Sequence''' will show you the protein FASTA sequence (translated from the DNA sequence) of the feature. | ||
| + | |||
| + | '''Feature Evidence vs. FIG''' and '''Feature Evidence vs. all DB''' link to the [[SEED_Viewer_Manual/Evidence|Evidence]] page. The difference between the two is that the evidence shown for the feature includes only evidence against features in the SEED or also against other databases (e.g. GenBank, SwissProt, UniProt and many others). The evidence page will also allow you to change this selection. | ||
| + | |||
| + | [[Image:MenuFeature.png]] | ||
| + | |||
| + | === Feature Tools === | ||
| + | |||
| + | This menu is also present if a feature is defined. It lets the user run a variaty of tools using the feature sequence. These include tools that look for transmembrane helices (e.g. [http://www.cbs.dtu.dk/services/TMHMM/ TMHMM]), signal peptides ([http://www.psort.org/ PSORT], [http://www.cbs.dtu.dk/services/SignalP/ SignalP]), protein domains (e.g. [http://www.ebi.ac.uk/interpro/ InterPro], [http://prodom.prabi.fr/prodom/current/html/home.php ProDom]) and others. | ||
| + | |||
| + | [[Image:MenuFeatTools.png]] | ||
| + | |||
| + | === FIGfams Menu === | ||
| + | |||
| + | On SeedViewer pages that deal with [[Glossary#FIGfam|FIGfams]], you will find an additional FIGfams menu. The '''Home''' link leads to the entry page for [[SEED_Viewer_Manual/FIGfams|FIGfams]]. | ||
| + | |||
| + | The '''Release History''' will lead you to a [[SEED_Viewer_Manual/FIGfamsRelease|page]] containing a number of statistics about the current release of the FIGfams. | ||
| + | |||
| + | '''Download''' will connect you to the FTP server to enable you to download the FIGfams. | ||
| + | |||
| + | [[Image:MenuFIGfam.png]] | ||
Latest revision as of 04:25, 5 December 2008
Menu Overview
The menu is a small bar below the logo of each page. Most menus have submenus that can be accessed via hovering over the menu. Click the submenu entry you want to go to. If no submenu is present, you can directly click the menu itself.
This menu is present on every SeedViewer page. The first entry Startpage will lead to the SeedViewer HomePage.
Organisms will open the Organism Select page. You will see an overview of all organisms in the SEED and are able to select one to view in detail.
Subsystems will do the same for subsystems.
Scenarios lets you browse Scenarios on the Scenarios Page.
FIGFams leads to an entry point for browsing FIGfams. You can find more information about FIGfams here.
The BLAST Search links to the BLAST Page. You will be able to BLAST a sequence against an organism in the SEED.
Help Menu
This menu is also present on all SeedViewer pages. The first link What is the SEED will lead you to the SEED Homepage.
Clicking the second link How to use the SEED Viewer will get you to this wiki.
Submitting data to SEED lets you browse the RAST Tutorial.
Contact enables you to write an email to the SEED team.
Clicking Register is the first step to get a user account. It will lead to the Register page.
I forgot my password enables you to Request a new password.
Organism Menu
In the context of a selected organism, an organism menu will appear. General Information will lead to the Organism Page of the selected organism.
The Feature Table will open the Genome Browser and show you the features present in that organism.
Genome Browser leads to the Genome Browser for the selected organism.
Clicking Scenarios shows the Scenarios page for your organism.
Subsystems opens a page that lets you select subsystems.
Export enables you to download the features of your organism.
Comparative Tools
This menu is also present whenever an organism is viewed. Different kinds of comparisons of your selected organism to other organisms are available here.
Function based Comparison - The Compare Metabolic Reconstruction will enable you to see the metabolic reconstruction of your selected organism against that of another one.
The Sequence based Comparison Multi Genome Compare shows a table and a graphic comparing a selected set of organisms projected against your chosen one (BLAST-based).
Use the KEGG Metabolic Analysis to project the metabolic capabilities of your organism onto KEGG maps.
Blasting against your organism is possible using the BLAST Search.
Feature Menu
Whenever a feature is defined on a SeedViewer page, you will find the Feature menu for that feature.
The Feature Overview points to the Annotation page. It shows general information about the feature, als well as a Compare Regions View that displays the feature in its genomic context and in comparison to homologs in other genomes.
DNA Sequence will open a page with the DNA Sequence of the feature (in FASTA format). DNA w/ flanking not only prints the DNA sequence of the feature, but also includes a user-defined number of bases upstream and downstream of the feature. Protein Sequence will show you the protein FASTA sequence (translated from the DNA sequence) of the feature.
Feature Evidence vs. FIG and Feature Evidence vs. all DB link to the Evidence page. The difference between the two is that the evidence shown for the feature includes only evidence against features in the SEED or also against other databases (e.g. GenBank, SwissProt, UniProt and many others). The evidence page will also allow you to change this selection.
Feature Tools
This menu is also present if a feature is defined. It lets the user run a variaty of tools using the feature sequence. These include tools that look for transmembrane helices (e.g. TMHMM), signal peptides (PSORT, SignalP), protein domains (e.g. InterPro, ProDom) and others.
FIGfams Menu
On SeedViewer pages that deal with FIGfams, you will find an additional FIGfams menu. The Home link leads to the entry page for FIGfams.
The Release History will lead you to a page containing a number of statistics about the current release of the FIGfams.
Download will connect you to the FTP server to enable you to download the FIGfams.