Difference between revisions of "SEED Viewer Manual/OrganismPage"
| (14 intermediate revisions by 2 users not shown) | |||
| Line 3: | Line 3: | ||
=== Menu and General Information === | === Menu and General Information === | ||
| − | Two | + | Two categories are added to the menu bar when visiting an organism page. These are organism specific menus described in the [[SEED_Viewer_Manual/Menu|Menu Overview]]. |
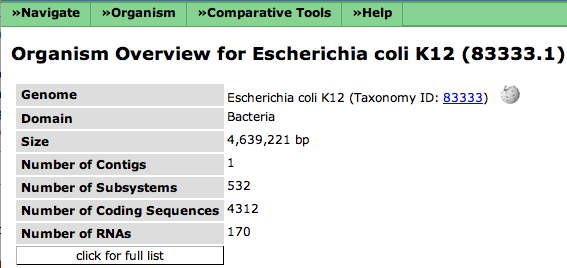
| − | The general information about an organism include name, taxonomy id (linked to NCBI), the domain (Bacteria, Archeae or Eukaryota) as well as some information about the genome (contigs, subsystems, genes). If you click on '''click for full list''', you will find more specific information about the organism. | + | The general information about an organism include name, taxonomy id (linked to NCBI), the domain (Bacteria, Archeae or Eukaryota) as well as some information about the genome (contigs, subsystems, genes). The icon next to the taxonomy id leads to a Wikipedia page for that organism (the icon only shows up if there is an existing Wikipedia page). |
| + | |||
| + | If you click on '''click for full list''', you will find more specific information about the organism. | ||
[[Image:OrgsMenuGeneral.png]] | [[Image:OrgsMenuGeneral.png]] | ||
| + | |||
| + | === Browse, Compare and Download === | ||
| + | |||
| + | The little [[WebComponents/Tabview|TabView]] next to the general information offers links to browse, compare and download the organism. These function are also available using the Organism menu. | ||
| + | |||
| + | '''Browse''' | ||
| + | |||
| + | Clicking the '''here''' link will lead to the [[SEED_Viewer_Manual/GenomeBrowser|Genome Browser]] for the selected organism. | ||
| + | It lets you browse the features of your organism. | ||
| + | |||
| + | [[Image:OrgsBrowse.png]] | ||
| + | |||
| + | '''Compare''' | ||
| + | |||
| + | Different kinds of comparisons of your selected organism to other organisms are available here (the function-based [[SEED_Viewer_Manual/CompareMetabolicReconstruction|Compare Metabolic Reconstruction]] will enable you to see the metabolic reconstruction of your selected organism against that of another one. The sequence-based [[SEED_Viewer_Manual/MultiGenomeCompare|Multi Genome Compare]] shows a table and a graphics comparing a selected set of organisms projected against your chosen one (BLAST-based). To project the metabolic capabilities of your organism on KEGG maps, use link to the [[SEED_Viewer_Manual/KEGG|KEGG]] page. Blasting against your organism is enabled using the [[SEED_Viewer_Manual/BLASTOrganism|BLAST]] link. | ||
| + | |||
| + | [[Image:OrgsCompare.png]] | ||
| + | |||
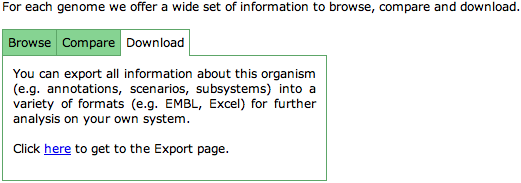
| + | '''Download''' | ||
| + | |||
| + | This tab provides a link to the [[SEED_Viewer_Manual/DownloadOrganism|Download Organism]] page. | ||
| + | |||
| + | [[Image:OrgsDownload.png]] | ||
| + | |||
| + | === Subsystem Information === | ||
| + | |||
| + | This part of the Organism Page deals with the subsystems present in your organism. The information is organized in a tab view, where the first tab shows a graphical overview of the subsystem information, while the second part lists all features in subsystems in a [[WebComponents/Table|Table]]. | ||
| + | |||
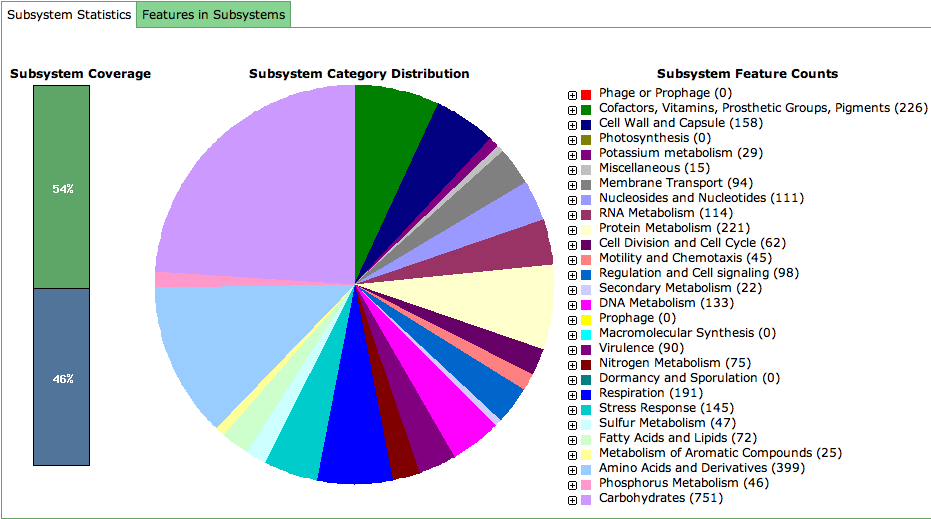
| + | '''(1) Subsystem Statistics''' | ||
| + | |||
| + | The leftmost bar chart (Subsystem Coverage) depicts the percentage of features from the selected organism that are in subsystems. | ||
| + | |||
| + | In the middle the pie chart shows the distribution of subsystem categories in the organism. Hovering over the slices will inform you about the category you're looking at and the number of genes that fall in that category. | ||
| + | |||
| + | Right to the pie chart, you can find the Subsystem Feature Counts. The slices in the pie chart are presented in a tree view. Clicking the '''+''' signs will open a category and show the subcategory. Clicking the '''+''' of a subcategory will show the subsystems in that subcategory that are present in the organism. The links of the subsystems will open a [[SEED_Viewer_Manual/Subsystems|Subsystems Page]] for that subsystem. Behind all (sub)categories and subsystem you can find the number of genes that are present in the subsystem for that organism. | ||
| + | |||
| + | [[Image:OrgsSubsystems.png]] | ||
| + | |||
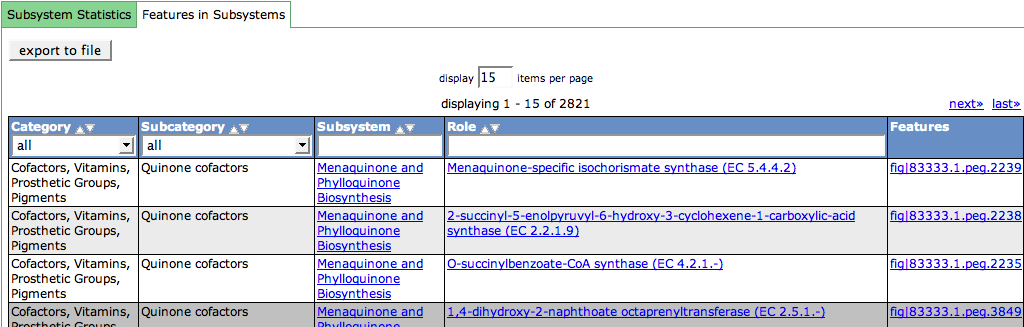
| + | '''(2) Features in Subsystems''' | ||
| + | |||
| + | Click on the second tab of the tab view to get the tabular view of the Features in Subsystems. | ||
| + | The [[WebComponents/Table|Table]] shows all the Subsystem Category, Subcategory, Subsystem Name, the Functional Role and the Feature Name of all features in subsystems in the organism. | ||
| + | |||
| + | The Subsystem links again lead to the [[SEED_Viewer_Manual/Subsystems|Subsystems]] page. Functional Role links will show the [[SEED_Viewer_Manual/FunctionalRoles|Functional Role]] page for that role. Feature links go to the [[SEED_Viewer_Manual/Annotation|Annotation page]] for that feature. | ||
| + | |||
| + | [[Image:OrgsFeatures.png]] | ||
Latest revision as of 08:48, 18 November 2008
Organism Page
Menu and General Information
Two categories are added to the menu bar when visiting an organism page. These are organism specific menus described in the Menu Overview.
The general information about an organism include name, taxonomy id (linked to NCBI), the domain (Bacteria, Archeae or Eukaryota) as well as some information about the genome (contigs, subsystems, genes). The icon next to the taxonomy id leads to a Wikipedia page for that organism (the icon only shows up if there is an existing Wikipedia page).
If you click on click for full list, you will find more specific information about the organism.
Browse, Compare and Download
The little TabView next to the general information offers links to browse, compare and download the organism. These function are also available using the Organism menu.
Browse
Clicking the here link will lead to the Genome Browser for the selected organism. It lets you browse the features of your organism.
Compare
Different kinds of comparisons of your selected organism to other organisms are available here (the function-based Compare Metabolic Reconstruction will enable you to see the metabolic reconstruction of your selected organism against that of another one. The sequence-based Multi Genome Compare shows a table and a graphics comparing a selected set of organisms projected against your chosen one (BLAST-based). To project the metabolic capabilities of your organism on KEGG maps, use link to the KEGG page. Blasting against your organism is enabled using the BLAST link.
Download
This tab provides a link to the Download Organism page.
Subsystem Information
This part of the Organism Page deals with the subsystems present in your organism. The information is organized in a tab view, where the first tab shows a graphical overview of the subsystem information, while the second part lists all features in subsystems in a Table.
(1) Subsystem Statistics
The leftmost bar chart (Subsystem Coverage) depicts the percentage of features from the selected organism that are in subsystems.
In the middle the pie chart shows the distribution of subsystem categories in the organism. Hovering over the slices will inform you about the category you're looking at and the number of genes that fall in that category.
Right to the pie chart, you can find the Subsystem Feature Counts. The slices in the pie chart are presented in a tree view. Clicking the + signs will open a category and show the subcategory. Clicking the + of a subcategory will show the subsystems in that subcategory that are present in the organism. The links of the subsystems will open a Subsystems Page for that subsystem. Behind all (sub)categories and subsystem you can find the number of genes that are present in the subsystem for that organism.
(2) Features in Subsystems
Click on the second tab of the tab view to get the tabular view of the Features in Subsystems. The Table shows all the Subsystem Category, Subcategory, Subsystem Name, the Functional Role and the Feature Name of all features in subsystems in the organism.
The Subsystem links again lead to the Subsystems page. Functional Role links will show the Functional Role page for that role. Feature links go to the Annotation page for that feature.