SEED Viewer Manual/Editing Capabilities/CreateFeature
Editing Capabilities - Create Feature
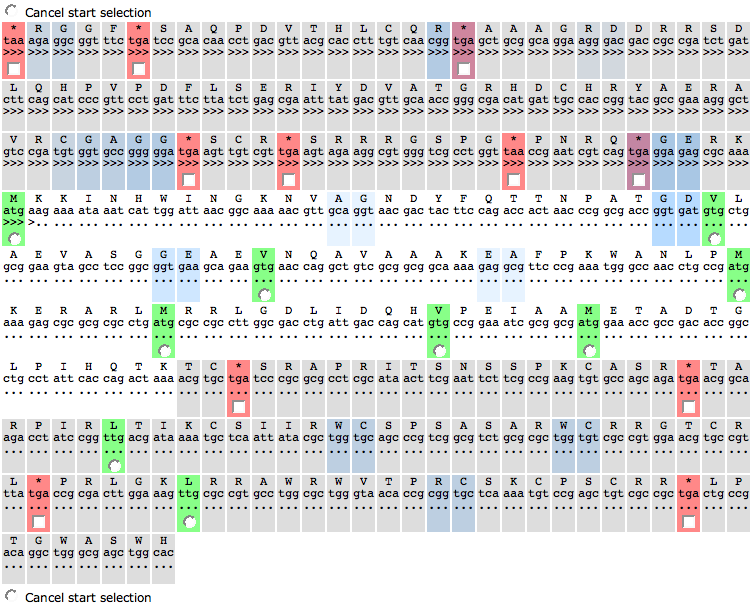
This page lets you create a new protein feature. It consists of three parts, a Control TabView, an Actions panel and the Sequence Table.
Control TabView
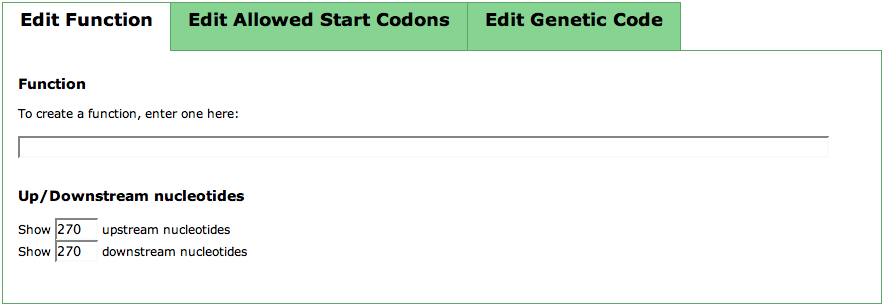
In the first tab of this TabView, you can put in a function for your newly created feature by just entering it into the text field. Below that, you can see two text fields for controlling Up/Downstream Nucleotides'. As you usually enter this page from the Search Gene Page, you have a hit region defined in which the feature is to be located. The numbers you can enter are the number of upstream and downstream nucleotides of the hit you want to see in the Sequence Table. After entering your numbers, click the update button in the Actions panel
The second tab of the TabView lists all codons of the genetic code. Here, you can select the start codons that are possible for the genome you want to create a feature in. Pre-selected are the standard start codons that are usually used for bacterial genomes. You can choose a different set of codons and press the button update in the Actions panel to change the view in the Sequence Table.
The genetic code can have small differences from the default genetic code in some organisms. You can change the standard genetic code in the third tab of the TabView. For each triplet, you can see the default amino acid that is usually used in organisms. If you want to change this for a triplet, click the respective drop down box and choose the right amino acid. To make the changes visible in the Sequence Table, press the update button in the Actions panel again.