Difference between revisions of "SEED Viewer Manual/Editing Capabilities/ChromosomalClusters"
| Line 20: | Line 20: | ||
Editing the function for the features in the Details table is done by checking a function or entering a new one, then clicking the '''annotate''' button. You can select a UniProt function, or one of the other feature's functions. If you want to use a new one, enter it in the '''new function''' text field and check the radio box next to it. The new function will be used to annotate all features that are checked in the '''Feature''' column. You will only be able to check features you can edit. In this example, only the ''Escherichia coli K12'' can be edited, so you will find a checkbox only for this feature. | Editing the function for the features in the Details table is done by checking a function or entering a new one, then clicking the '''annotate''' button. You can select a UniProt function, or one of the other feature's functions. If you want to use a new one, enter it in the '''new function''' text field and check the radio box next to it. The new function will be used to annotate all features that are checked in the '''Feature''' column. You will only be able to check features you can edit. In this example, only the ''Escherichia coli K12'' can be edited, so you will find a checkbox only for this feature. | ||
| + | |||
| + | To align features in the [[SEED_Viewer_Manual/AlignSeqs|Alignment Page]], check the features of interest and press the button '''align'''. | ||
| + | |||
| + | The buttons '''check all''', '''uncheck all''' and '''check to last checked''' can be used for the check boxes of the features, which is especially important for large tables. | ||
[[Image:Chromosomal2.png]] | [[Image:Chromosomal2.png]] | ||
Latest revision as of 07:26, 8 December 2008
Editing Capabilities - Chromosomal Clusters
This page can be accessed from a Chromosomal Clusters view, using the button annotate clusters (only visible if you have editing rights for the genome).
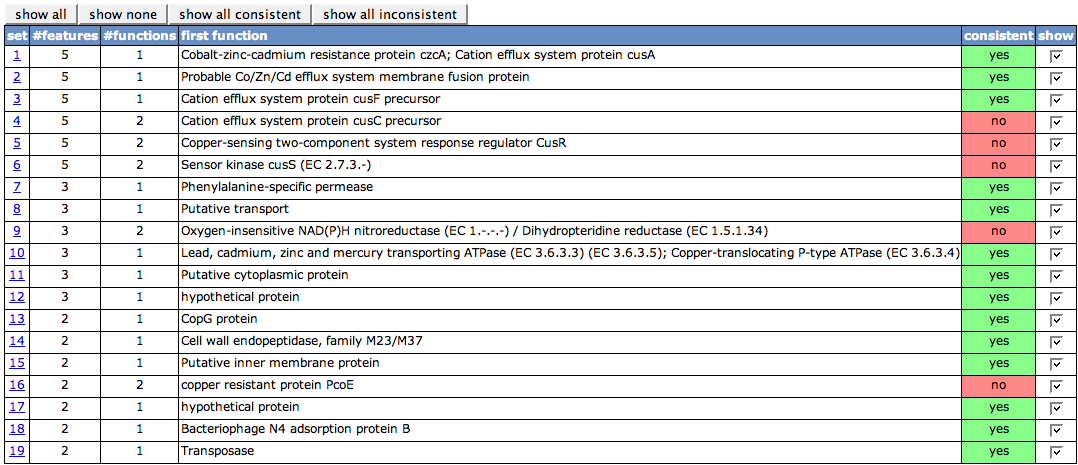
Overview Table
The first table on the page lists all sets of protein visible in the Chromosomal Clusters view. The set numbers in the first column are the set numbers from the view. The second column (#features) shows the number of features in the set. #functions depicts how many different functions the features in the set have. One of the functions is displayed in the column first function. Whenever there is more than one function, the cell in the next column will be painted in red, as it is not consistent, else green. With the check boxes in the last column, you can turn on and off the details tables for each of the sets.
Above the table, there are four buttons to turn on/off the details tables. show all will remove previous filtering and show you all details tables again. show none will remove all details tables, so that you can afterwards select single ones you'd like to see. show consistenct and show inconsistent is related to the consistent column in the table. It will display the details tables with a consistent or an inconsistent set, respectively.
Details table
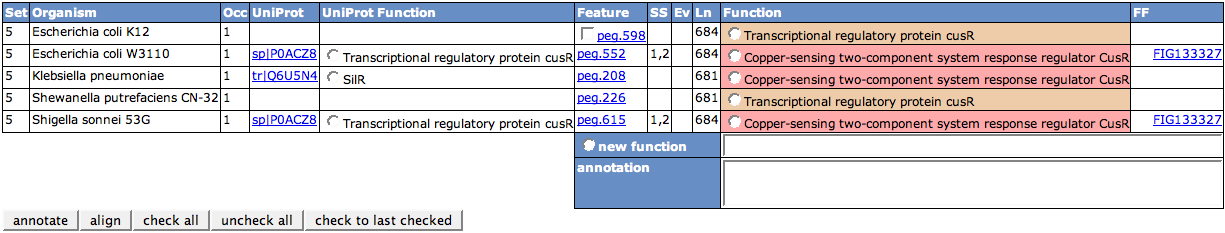
As a default, for each set you will get a details table in the following. These list all the features in the set. The set number is displayed in the first column. Organism is the genome the feature stems from. If there are more than one occurrence of features in the set for one genome, they are numbered in the Occ column. If there is a UniProt alias for the feature, the ID and the function of this alias will be shown in the next two columns.
The IDs of the features in the next column Feature link to the Annotation Page for that feature. If the feature is in a subsystem, you will find one or many (comma-separated) numbers in the SS column. Hovering over the cell will display a tooltip with the names of the subsystems. The column Ev shows Evidence Codes for the feature. There are three Evidence Codes that can be found in the last column. ISU means that the feature is unique in a cell of a subsystem. This means that there is no other feature in the genome that is thought to have the same function. ICW(number) means the feature is clustered with number features in the genome. FF says that it is in a FIGfam.
The length of the feature is shown in the next column (Ln). Its current function can be found in the column Function. Same colors in this column indicate same functions. Links to a FIGfam a feature is member of are displayed in the last column.
Editing the function for the features in the Details table is done by checking a function or entering a new one, then clicking the annotate button. You can select a UniProt function, or one of the other feature's functions. If you want to use a new one, enter it in the new function text field and check the radio box next to it. The new function will be used to annotate all features that are checked in the Feature column. You will only be able to check features you can edit. In this example, only the Escherichia coli K12 can be edited, so you will find a checkbox only for this feature.
To align features in the Alignment Page, check the features of interest and press the button align.
The buttons check all, uncheck all and check to last checked can be used for the check boxes of the features, which is especially important for large tables.