Difference between revisions of "SEED Viewer Manual/CompareMetabolicReconstruction"
| (4 intermediate revisions by the same user not shown) | |||
| Line 3: | Line 3: | ||
This comparison is done on basis of the functions of all features of two genomes. The reference genome is called '''Organism A''', the comparison genome '''Organism B'''. | This comparison is done on basis of the functions of all features of two genomes. The reference genome is called '''Organism A''', the comparison genome '''Organism B'''. | ||
| − | + | Entering the page, you have already defined an '''Organism A''', as you followed a link e.g. from an [[SEED_Viewer_Manual/OrganismPage|Organism Page]] of the '''Organism A'''. Now you have to choose your '''Organism B''' using the [[SEED_Viewer_Manual/OrganismSelect|Organism Select]] on the page. Click '''select''' to start the computation. | |
| − | The | + | The resulting page will start with a header line showing the two genomes to compare. Behind the header, you find a button '''select other''' if you don't like the comparison genome. It will open an [[SEED_Viewer_Manual/OrganismSelect|Organism Select]] to choose another comparison organism. |
| − | + | [[Image:CompareMetaResult.png]] | |
| − | [[Image:CompareMetaReconst]] | + | The first column of the [[WebComponents/Table|table]] ('''Presence''') tells you for each function, if it is present in ''A'', ''B'', or ''A and B''. The next three columns ('''Category''', '''Subcategory''' and '''Subsystem''') describe the subsystem the functional role belongs to. The functional role itself is displayed in column 5 ('''Role'''). |
| + | |||
| + | The feature(s) that implement(s) the functional role in '''Organism A''' will appear in the next column. They are linked to the [[SEED_Viewer_Manual/Annotation|Annotation Page]] for that feature. The following column '''SS active A''' tells you if the subsystem listed in the line is active in the Organism A, meaning that it has a valid variant code. The same information is shown for '''Organism B''' in the next two columns. | ||
| + | |||
| + | If a role is not present for one of the organisms, a '''Find''' button will be displayed instead of the feature id. It leads to the [[SEED_Viewer_Manual/SearchGene|Search Gene Page]]. Here, you can search for a candidate for the function. | ||
| + | |||
| + | If a feature is assigned with more than one functional role (multi-functional), it will appear once for each function in the table. | ||
| + | |||
| + | You can export the contents of the table in ''tab-separated format'' using the '''save to file''' button. | ||
| + | |||
| + | [[Image:CompareMetaReconst.png]] | ||
Latest revision as of 08:43, 2 December 2008
Compare Metabolic Reconstruction
This comparison is done on basis of the functions of all features of two genomes. The reference genome is called Organism A, the comparison genome Organism B.
Entering the page, you have already defined an Organism A, as you followed a link e.g. from an Organism Page of the Organism A. Now you have to choose your Organism B using the Organism Select on the page. Click select to start the computation.
The resulting page will start with a header line showing the two genomes to compare. Behind the header, you find a button select other if you don't like the comparison genome. It will open an Organism Select to choose another comparison organism.
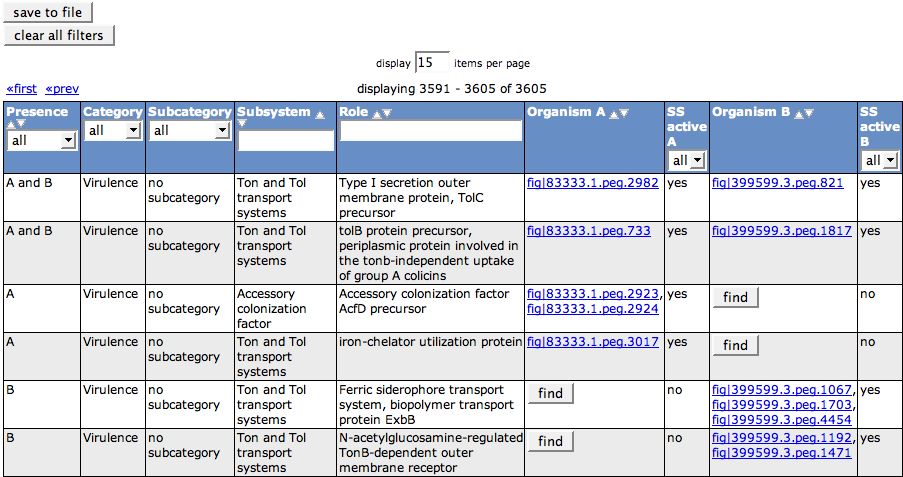
The first column of the table (Presence) tells you for each function, if it is present in A, B, or A and B. The next three columns (Category, Subcategory and Subsystem) describe the subsystem the functional role belongs to. The functional role itself is displayed in column 5 (Role).
The feature(s) that implement(s) the functional role in Organism A will appear in the next column. They are linked to the Annotation Page for that feature. The following column SS active A tells you if the subsystem listed in the line is active in the Organism A, meaning that it has a valid variant code. The same information is shown for Organism B in the next two columns.
If a role is not present for one of the organisms, a Find button will be displayed instead of the feature id. It leads to the Search Gene Page. Here, you can search for a candidate for the function.
If a feature is assigned with more than one functional role (multi-functional), it will appear once for each function in the table.
You can export the contents of the table in tab-separated format using the save to file button.