Difference between revisions of "SEED Viewer Manual/Annotation"
| Line 20: | Line 20: | ||
'''FIGfams''' are protein families based on the subsystems technology. If you find an entry in this fields, the feature is part of the stated FIGfam. The link leads to the [[SEED_Viewer_Manual/FIGfams|FIGfam Viewer]]. | '''FIGfams''' are protein families based on the subsystems technology. If you find an entry in this fields, the feature is part of the stated FIGfam. The link leads to the [[SEED_Viewer_Manual/FIGfams|FIGfam Viewer]]. | ||
| + | |||
| + | '''database cross references''' link the feature to its entries in other databases like [[www.uniprot.org|UniProt]], | ||
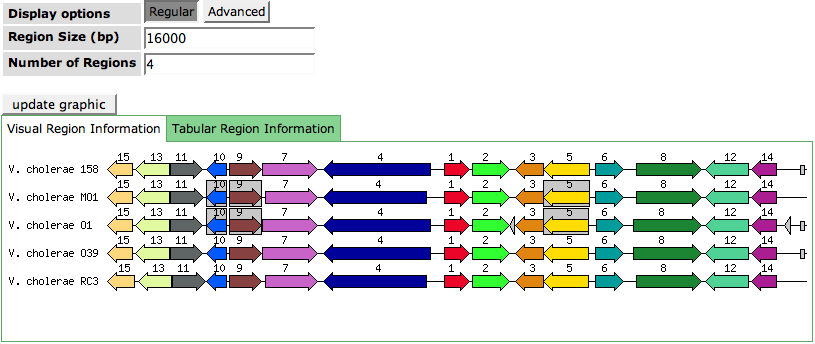
[[Image:AnnotationFeat.png]] | [[Image:AnnotationFeat.png]] | ||
Revision as of 08:06, 21 November 2008
Annotation
The Annotation page shows a variaty of information about a single feature like a protein or an RNA. The page is roughly divided into three parts. The Annotation Overview presents the basic information about the feature. Reasons for Current Assignment reflect why the feature was assigned with the current functional role. The third part is a Compare Regions View showing the region of the feature in context to its own and related genomes.
If you are logged in and the feature belongs to your private genome, this page will have additional options for you to annotate the feature. These are described here.
The Annotation Overview
The feature ID and the genome it belongs to are shown in the header line of this part of the page. They link to Genome Browser and the Organism Page, respectively.
The current annotation depicts the functional role that is currently assigned to the feature. As annotations can be changed by users, you have the option to view an annotation history by pressing the show button in the cell annotation history some rows below. It will open a small table listing the date, the curator and the annotation that was made for each entry.
As the genome name for the feature is already presented in the header of this section, we additionally show the taxonomy id for that genome in the overview. The link will lead to the Taxonomy Browser at the NCBI showing the taxonomy information for that genome.
The internal links you can see in the next row are leading to different pages containing other views and information for the feature. The Genome Browser (genome browser) displays the feature in the context of its genome. evidence leads to the Evidence page showing evidence for the annotation of the feature in form of Similarities and protein domains. Downloading the actual sequence for the feature is possible using the Sequence Page (sequence) link.
At PubMed Links you can see the PubMed IDs to papers linked to the NCBI Entrez Database. The PubMed IDs shown are direct literature links attached directly to a feature, so-called dlits.
FIGfams are protein families based on the subsystems technology. If you find an entry in this fields, the feature is part of the stated FIGfam. The link leads to the FIGfam Viewer.
database cross references link the feature to its entries in other databases like UniProt,