Difference between revisions of "SEED Viewer Manual/BLASTOrganism"
| Line 1: | Line 1: | ||
| + | == Blast against an organism == | ||
| + | |||
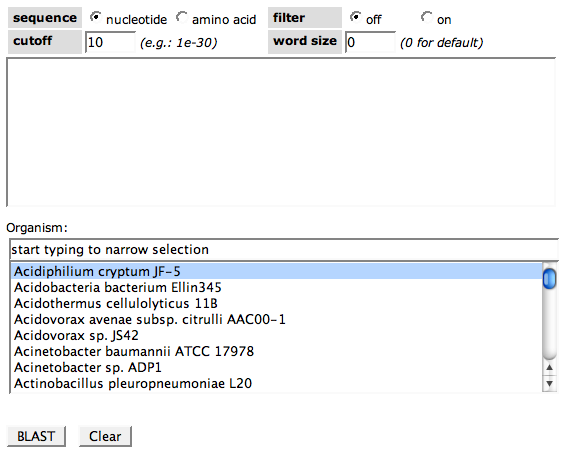
| + | Blasting a sequence against an organism includes two steps. First, you paste your custom sequence into the first text field. | ||
| + | |||
| + | Choose nucleotide or amino acid depending on your input sequence (DNA or protein) in the '''sequence''' cell. The BLAST filter can be turned on or off in the '''filter''' cell. Choose the evalue '''cutoff''' you want to apply to the results (Hits with a larger evalue will not be shown). The '''word size''', a BLAST parameter for building the seeds of hits, can also be changed if needed. | ||
| + | |||
| + | As a second step, choose the genome you want to BLAST against in the select box. | ||
| + | |||
| + | Click the button '''BLAST''' if you are ready. '''Clear''' will reset all options. | ||
| + | |||
| + | The output of the BLAST search will provide links of the hits into the [[SEED_Viewer_Manual/GenomeBrowser|Genome Browser]]. The region that is hit in the genome will be marked using a red box. | ||
| + | |||
[[Image:BlastOrg.png]] | [[Image:BlastOrg.png]] | ||
Latest revision as of 10:42, 18 November 2008
Blast against an organism
Blasting a sequence against an organism includes two steps. First, you paste your custom sequence into the first text field.
Choose nucleotide or amino acid depending on your input sequence (DNA or protein) in the sequence cell. The BLAST filter can be turned on or off in the filter cell. Choose the evalue cutoff you want to apply to the results (Hits with a larger evalue will not be shown). The word size, a BLAST parameter for building the seeds of hits, can also be changed if needed.
As a second step, choose the genome you want to BLAST against in the select box.
Click the button BLAST if you are ready. Clear will reset all options.
The output of the BLAST search will provide links of the hits into the Genome Browser. The region that is hit in the genome will be marked using a red box.