Difference between revisions of "MG RAST v2.0"
FolkerMeyer (talk | contribs) m |
FolkerMeyer (talk | contribs) |
||
| (15 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
== MG-RAST v2.0 Homepage == | == MG-RAST v2.0 Homepage == | ||
| − | === What is MG-RAST === | + | === What is MG-RAST ( Executive Summary) === |
MG-RAST (Metagenome Rapid Annotation using Subsystem Technology) is a fully-automated service for annotating metagenome samples. It is the only service that we are aware of that will handle both short reads (notably reads of 109 or 230 basepairs from a [http://en.wikipedia.org/wiki/454_life_sciences#Technology 454] instrument) and longer [http://en.wikipedia.org/wiki/DNA_sequencing#Chain-termination_methods Sanger] reads or even assembled short contigs. | MG-RAST (Metagenome Rapid Annotation using Subsystem Technology) is a fully-automated service for annotating metagenome samples. It is the only service that we are aware of that will handle both short reads (notably reads of 109 or 230 basepairs from a [http://en.wikipedia.org/wiki/454_life_sciences#Technology 454] instrument) and longer [http://en.wikipedia.org/wiki/DNA_sequencing#Chain-termination_methods Sanger] reads or even assembled short contigs. | ||
| Line 7: | Line 7: | ||
In the MG-RAST analysis the fragments in a given sample will be compared to protein, RNA and subsystem databases. | In the MG-RAST analysis the fragments in a given sample will be compared to protein, RNA and subsystem databases. | ||
| + | Both the submitted data and the MG-RAST analysis can be shared with other users or "published" to the public on the server, we provide stable unique identifiers for public metagenomes. | ||
=== Overview === | === Overview === | ||
| Line 19: | Line 20: | ||
| − | === What's new? (Changes from MG-RAST v1.2 | + | === What's new in v2.0? (Changes from MG-RAST v1.2) === |
We have gone through several rounds of feedback with users of version 1.2 (Many thanks to all who send suggestions!) and have included the following new capabilities in version 2.0: | We have gone through several rounds of feedback with users of version 1.2 (Many thanks to all who send suggestions!) and have included the following new capabilities in version 2.0: | ||
* Significant improvements of user interface responsiveness and overall performance. | * Significant improvements of user interface responsiveness and overall performance. | ||
| − | * | + | * The ability to <strong>"publish" metagenomes on the MG-RAST server</strong> for public use. |
| − | * The ability to download subsets of fragments as fasta (eg. all fragments matching a given [http://www.theseed.org/wiki/Glossary#Subsystem subsystem] or [http://www.theseed.org/wiki/Glossary#Functional_role functional role]). | + | * The ability to <strong>download subsets</strong> of fragments as fasta (eg. all fragments matching a given eg. a [http://www.theseed.org/wiki/Glossary#Subsystem subsystem] or a [http://www.theseed.org/wiki/Glossary#Functional_role functional role]). |
| − | * The ability to modify parameters for sequence comparison underlying both metabolic reconstruction and phylogenetic reconstruction on the fly. | + | * The <strong>ability to modify parameters for sequence comparison</strong> underlying both metabolic reconstruction and phylogenetic reconstruction on the fly. |
* The same capability for the heatmap style comparisons of both metabolisms and phylogenetic reconstructions. | * The same capability for the heatmap style comparisons of both metabolisms and phylogenetic reconstructions. | ||
| − | * We have added a recruitment plot feature, plotting fragments against microbial genomes. | + | * We have added a <strong> recruitment plot </strong>feature, plotting fragments against microbial genomes. |
| + | * We have added the ability to <strong> view all BLAST hits for a fragment </strong> and show the individual BLAST alignments. | ||
* The ability to use [http://www.genome.ad.jp/kegg/kegg2.html KEGG] maps map explore and compare metabolic reconstructions on several hierarchy levels (e.g. the high level metabolism overview). | * The ability to use [http://www.genome.ad.jp/kegg/kegg2.html KEGG] maps map explore and compare metabolic reconstructions on several hierarchy levels (e.g. the high level metabolism overview). | ||
* We have changed the pipeline that computes the underlying data so all numbers/percentages/comparisons/etc. will have changed if you look at your data in v2.0 | * We have changed the pipeline that computes the underlying data so all numbers/percentages/comparisons/etc. will have changed if you look at your data in v2.0 | ||
| − | * We have also updated the underlying databases. Most notably the [http://www.theseed.org SEED] NR no longer from represents the status from 2006, we have added the [http://www.arb-silva.de/ Silva RNA database]) | + | * We have also <strong>updated the underlying databases</strong>. Most notably the [http://www.theseed.org SEED] NR no longer from represents the status from 2006, we have added the [http://www.arb-silva.de/ Silva RNA database]) |
| − | * Invite a friend feature to share data that you submitted with other users by just entering their email addresses. | + | * Added the <strong>Invite a friend feature</strong> to share data that you submitted with other users by just entering their email addresses. |
* Support for user driven creation and maintenance of groups. | * Support for user driven creation and maintenance of groups. | ||
| − | * | + | * The ability to support arbitrary sets and versions of databases. |
| + | * Many small detailed fixes and improvements. | ||
| − | + | == Databases used == | |
* [http://www.theseed.org SEED non redundant protein database] | * [http://www.theseed.org SEED non redundant protein database] | ||
* [http://greengenes.lbl.gov/ GREENGENES] | * [http://greengenes.lbl.gov/ GREENGENES] | ||
| Line 41: | Line 44: | ||
* [http://www.arb-silva.de/ SILVA] | * [http://www.arb-silva.de/ SILVA] | ||
| − | + | == Standard Compliance == | |
We are currently working with Dawn Fields and Renzo Kottmann from the GSC to support a version of "ANDREAS PLEASE SUPPLY THIS INFO" | We are currently working with Dawn Fields and Renzo Kottmann from the GSC to support a version of "ANDREAS PLEASE SUPPLY THIS INFO" | ||
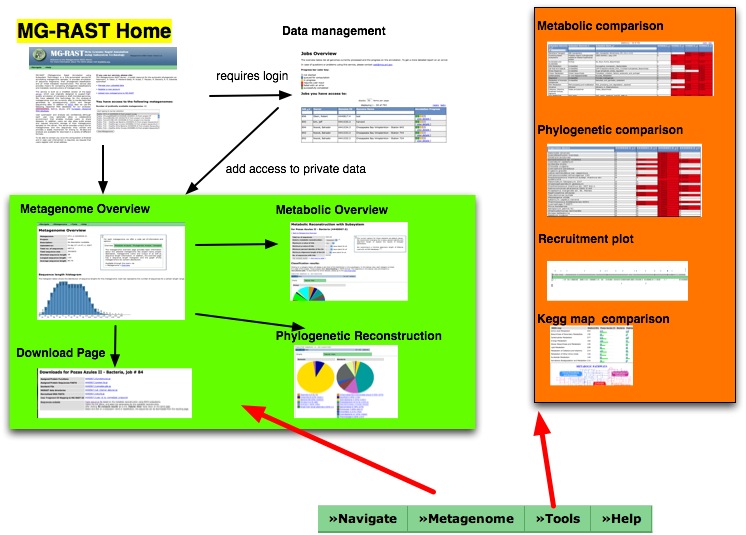
| − | == | + | == Navigating MG-RAST v2.0 == |
| − | + | [[Image:MG-Rast Navigation Overview.jpg]] | |
| − | + | ||
| − | + | == Manual for MG-RAST v2.0 == | |
| − | + | Separate page | |
| + | == Tutorial (Using MG-RAST-v2.0 to analyze my metagenome sample) == | ||
| + | A [[MG_RAST_v2.0_tutorial|tour]] of the user interface. | ||
| + | |||
| + | == Availability== | ||
| + | Using MG-RAST is free for all users. We provide privacy for the data submitted and | ||
| + | |||
MG-RAST is open source. While we currently do not provide a defined release, current snapshots of the system are available via CVS. | MG-RAST is open source. While we currently do not provide a defined release, current snapshots of the system are available via CVS. | ||
| − | === | + | == Frequently asked questions (FAQ) == |
| − | - | + | |
| + | We provide a list of frequently asked questions at [[MG_RAST_v2.0_FAQ]]. | ||
| + | |||
| + | == Contact == | ||
| + | Once logged in to the system, the address of the MG-RAST system will be available to all users. | ||
Latest revision as of 16:24, 28 June 2008
MG-RAST v2.0 Homepage
What is MG-RAST ( Executive Summary)
MG-RAST (Metagenome Rapid Annotation using Subsystem Technology) is a fully-automated service for annotating metagenome samples. It is the only service that we are aware of that will handle both short reads (notably reads of 109 or 230 basepairs from a 454 instrument) and longer Sanger reads or even assembled short contigs.
In the MG-RAST analysis the fragments in a given sample will be compared to protein, RNA and subsystem databases.
Both the submitted data and the MG-RAST analysis can be shared with other users or "published" to the public on the server, we provide stable unique identifiers for public metagenomes.
Overview
The MG-RAST system will utilize data structures and software generated in the context of The SEED and NMPDR to provide annotation of sequence fragments, their phylogenetic classification and an initial metabolic reconstruction. The service also provides means for comparing phylogenetic classifications and metabolic reconstructions of metagenomes.
The service is built as a modified version of the RAST server, which was originally designed to support high-quality annotation of complete or draft microbial genomes. We have adapted this technology for the analysis of metagenomes. In addition to SEED data we use the following ribosomal RNA databases for our analyses: GREENGENES, RDP-II, SILVA, and European ribosomal RNA database.
User submission and analysis are confidential, although each user may optionally allow a collaborative environment that enables multiple users to share datasets. In addition, users can also allow public access and request long-term storage of their metagenomic samples on the server. The server provides unique IDs for metagenomes and the sequences they contain and provides a stable mechanism for linking to. All data and analysis are available for download in a variety of different formats.
To be able to contact you once the computation is finished and in case user intervention is required, we request that users register with email address.
What's new in v2.0? (Changes from MG-RAST v1.2)
We have gone through several rounds of feedback with users of version 1.2 (Many thanks to all who send suggestions!) and have included the following new capabilities in version 2.0:
- Significant improvements of user interface responsiveness and overall performance.
- The ability to "publish" metagenomes on the MG-RAST server for public use.
- The ability to download subsets of fragments as fasta (eg. all fragments matching a given eg. a subsystem or a functional role).
- The ability to modify parameters for sequence comparison underlying both metabolic reconstruction and phylogenetic reconstruction on the fly.
- The same capability for the heatmap style comparisons of both metabolisms and phylogenetic reconstructions.
- We have added a recruitment plot feature, plotting fragments against microbial genomes.
- We have added the ability to view all BLAST hits for a fragment and show the individual BLAST alignments.
- The ability to use KEGG maps map explore and compare metabolic reconstructions on several hierarchy levels (e.g. the high level metabolism overview).
- We have changed the pipeline that computes the underlying data so all numbers/percentages/comparisons/etc. will have changed if you look at your data in v2.0
- We have also updated the underlying databases. Most notably the SEED NR no longer from represents the status from 2006, we have added the Silva RNA database)
- Added the Invite a friend feature to share data that you submitted with other users by just entering their email addresses.
- Support for user driven creation and maintenance of groups.
- The ability to support arbitrary sets and versions of databases.
- Many small detailed fixes and improvements.
Databases used
Standard Compliance
We are currently working with Dawn Fields and Renzo Kottmann from the GSC to support a version of "ANDREAS PLEASE SUPPLY THIS INFO"
Manual for MG-RAST v2.0
Separate page
Tutorial (Using MG-RAST-v2.0 to analyze my metagenome sample)
A tour of the user interface.
Availability
Using MG-RAST is free for all users. We provide privacy for the data submitted and
MG-RAST is open source. While we currently do not provide a defined release, current snapshots of the system are available via CVS.
Frequently asked questions (FAQ)
We provide a list of frequently asked questions at MG_RAST_v2.0_FAQ.
Contact
Once logged in to the system, the address of the MG-RAST system will be available to all users.