Difference between revisions of "SEED Viewer Tutorial"
| Line 10: | Line 10: | ||
[[Image:seed_fig1.jpg]] | [[Image:seed_fig1.jpg]] | ||
| + | |||
Figure 1. SEED Viewer navigation bar with links to the genome browser, genome features, reactions, and genome comparison tool. | Figure 1. SEED Viewer navigation bar with links to the genome browser, genome features, reactions, and genome comparison tool. | ||
Revision as of 22:32, 11 June 2007
The SEED Viewer
Selecting “Browse annotated genome in SEED Viewer” from the “Job Details” page of the RAST server will direct the user to the Organism Page in the SEED Viewer.
Organism Page in SEED Viewer
In the navigation bar at the top of the page adjacent to the homepage symbol is a pull down menu labeled 'This Organism' with the following 4 display options for this page:
Figure 1. SEED Viewer navigation bar with links to the genome browser, genome features, reactions, and genome comparison tool.
1. Overview and Subsystems
- The top of page provides summary statistics for this genome including the numbers of subsystems, coding sequences, RNAs and other background information.
- A breakdown of the number of protein encoding genes with hypothetical and non-hypothetical function assignments is provided in box in the top right corner of the page.
- Adjacent to the Taxonomy ID is the Wikipedia Globe that is linked to the Wikipedia entry for this organism.
- On the bottom left of the page a bar graph depicts the percentages of protein encoding genes for this genome that are present in at least one subsystem.
- Next to the bar graph a pie chart and accompanying expandable hierarchical tree, color coded to match the pie chart, organizes the subsystems present in this genome by cellular process. At every level in the tree the number of protein encoding genes in each category is listed in parentheses. At the leaves of the tree are links to a Subsytem page in the SEED Viewer.
2. Browse Genome
- This display allows you to walk the chromosome of the selected genome.
- If the genome is not completely assembled, the first contig will be selected.
- The window size in terms of number of base pairs can be selected using the 'Window Size' pulldown menu or the 'zoom in' or 'zoom out' buttons.
- The < and > buttons will move the window down or up the chromosome half the window size.
- The << and >> buttons will move the window down or up the chromosome a full window size.
- Under 'Options' you can select which features and the number of strands or reading frames displayed. By default the genome browser offers a two strand view of the currently selected organism.
- After selecting your options, click the 'Refresh' button to update the page.
- On the middle line of the chromosome display you will see features not associated with a reading frame, such as RNAs, pathogenicity islands or prophages.
- Hover over a feature in the chromosome display for a detailed description.
- Clicking an item will take you to the Annotation page in the SEED Viewer for this feature.
3. View Features
- Selecting this display provides a table containing feature ID, functional assignment, starting and stopping location in terms of base pairs relative to the beginning of the contig, feature type and a list of aliases.
- The total number of features for this genome is displayed above the table.
- Use the 'next>>' and 'last>>' links to quickly scroll through sets of 10 features.
- Filter features across the entire genome by using the text entry boxes in the 'Functional Assignment', 'Type' or 'Aliases' column header and then hitting enter.
- Sort or reverse sort the tables using the triangle in the columns headers.
- Export all or a subset of the features, after filtering, of a genome to a spreadsheet file using the 'Export Table' button.
4. View Reactions
- For each genome, the genes that are part of known subsystems are compiled as an expandable hierarchical tree. This organizes the subsystems present in this genome by cellular process.
- The next level in the tree shows all the reactions for a given cellular process with links to KEGG for complete reaction details.
- At the leaves of the tree are the fig ids for all proteins annotated as those which catalyze the reaction. These proteins are linked to an Annotation Overview in the SEED Viewer.
5. Compare Genome
This tool allows the user to identify protein functions that are the same or different for a set of organisms. The results are presented in tabular form with information regarding function, EC and subsystem involvement displayed. Each protein is linked to the Annotation Overview in the SEED Viewer.
6. Annotation Overview
- This page offers access to the data relating to a feature. By default the Overview version of the page is initially displayed providing the feature's function assignment, source organism, the role it performs in a subsystem, and a graphic displaying its genomic context. If this feature's function is associated with one or more functional roles, each functional role's name will be displayed and linked to a SEED Viewer Functional Role page. A part of a feature's function that is not also a functional role is NOT linked.
- If this feature is included in a subsystem, detailed information about this feature's role or roles in one or more subsystems is provided in a shaded box at the top of the page on the right. The functional role names and subsystem names are linked to the SEED Viewer Functional Role page and the SEED Viewer Subsystem page.
- In the graphic each line represents a different genome. The genome of the feature that is the subject of this page is on the first line with the subject feature in red and centered in a 16kb window. Features on the same line that have a gray shadow are functionally coupled to the subject feature. Features in different genome that share the same color or number are similar. Clicking 'Diverse Genomes' will change the genomes in the graphic to a set that is more phylogentically distant to the subject feature's genome.
- If there are no close genomes only a single line for the subject feature's genome will be present.
- If no data for diverse genomes has been computed a message saying so will be be displayed.
Sequence Data - From the green bar at the top of page, selecting the 'This Protein' menu will give the user the choice of viewing the this feature's DNA sequence, DNA with flanking sequence or amino acid sequence.
Tools - From the green bar at the top of the page, selecting the 'Tools' menu will give the user the choice of the following tools to analyze this feature:
- TMpred (transmembrane helix prediction)
- TMHMM (transmembrane helix prediction)
- Gram negative PSORT (subcellular localization prediction)
- Gram negative SignalP (signal sequence prediction)
- Gram positive PSORT (subcellular localization prediction)
- Gram positive SignalP (signal sequence prediction)
- LipoP (lipoprotein prediction)
- InterProScan (protein domain identification)
- Radar (detection of protein sequence repeats)
- PPSearch (protein motif identification)
- Gram negative CELLO (subcellular localization prediction)
- Gram positive CELLO (subcellular localization prediction)
- ProDom (protein domain identification)
Literature - This menu list links to sources of literature discussing this specific feature. Currently PubMed is the only source.
7. Functional Role
- This page displays statistics on the number of protein encoding genes within the SEED database that currently are associated with this functional role and within how many different organisms these genes are present.
- In addition, this page lists the subsystem(s) for a functional role and if present, EC number, associated GO terms, and KEGG reaction.
- A table lists all occurrences of the functional role in the SEED database.
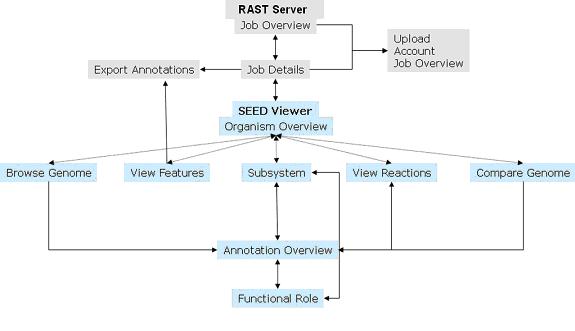
Figure 2. Flowchart for navigating the RAST Server and SEED Viewer. Arrows indicate how users can access the various pages within the system.