Difference between revisions of "SEED Viewer Manual/SearchGene"
| Line 13: | Line 13: | ||
In this case, a '''reference feature''' is used for a translated BLAST search against the whole genome sequence of the genome. On the top of the section, you can find a link to the used query feature. It leads to the [[SEED_Viewer_Manual/Annotation|Annotation Page]]. You can select a different query feature using the drop down box and click the button '''Take this gene for tblastn search'''. | In this case, a '''reference feature''' is used for a translated BLAST search against the whole genome sequence of the genome. On the top of the section, you can find a link to the used query feature. It leads to the [[SEED_Viewer_Manual/Annotation|Annotation Page]]. You can select a different query feature using the drop down box and click the button '''Take this gene for tblastn search'''. | ||
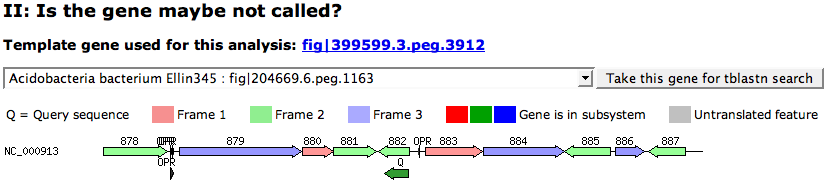
| − | The graphic shows the result of the tblastn search. It shows all found hit reagions. The hit is marked with a '''Q''' in the graphic. The region displays all features in the hit regions, colored by the frame each feature is in (See legend on top of the graphic). Dark colors mark that the features belong to the context subsystem. This way, you can see if the hit clusters | + | The graphic shows the result of the tblastn search. It shows all found hit reagions. The hit is marked with a '''Q''' in the graphic. The region displays all features in the hit regions, colored by the frame each feature is in (See legend on top of the graphic). Dark colors mark that the features belong to the context subsystem. This way, you can see if the hit clusters with other features of the subsystem. If the hit ('''Q''') overlaps with an existing feature in the same frame, this feature might be the candidate you look for. |
[[Image:SearchGene2.png]] | [[Image:SearchGene2.png]] | ||
Revision as of 11:17, 2 December 2008
Search Gene
In this page you can search for a function in a genome in the context of a subsystem. Two ways of looking for a candidate feature for the function are provided: Looking for already existing features in the genome using a protein-based BLAST search, and trying to find candidates via a translated BLAST search against the DNA sequence of the whole genome. In both cases, reference feature(s) are selected (or given to the page) from already existing features with assigned with the functional role that are part of the given subsystem.
Candidate genes found by Similarity (BLAST)?
This part looks for Similarities of all reference feature (features in the subsystem that are assigned with the function) to the features of the selected genome. The output table lists the E-Value (P-Sc), the reference feature that was hit (PEG), the length (Len) of the hit feature, the current function of the hit feature (Current fn), as well as the query feature (Matched peg), its length (Len), and its Function.
Is the gene maybe not called?
In this case, a reference feature is used for a translated BLAST search against the whole genome sequence of the genome. On the top of the section, you can find a link to the used query feature. It leads to the Annotation Page. You can select a different query feature using the drop down box and click the button Take this gene for tblastn search.
The graphic shows the result of the tblastn search. It shows all found hit reagions. The hit is marked with a Q in the graphic. The region displays all features in the hit regions, colored by the frame each feature is in (See legend on top of the graphic). Dark colors mark that the features belong to the context subsystem. This way, you can see if the hit clusters with other features of the subsystem. If the hit (Q) overlaps with an existing feature in the same frame, this feature might be the candidate you look for.