Difference between revisions of "SEED Viewer Manual/SearchGene"
| Line 1: | Line 1: | ||
== Search Gene == | == Search Gene == | ||
| − | In this page you can search for a function in a genome in the context of a subsystem. Two ways of looking for a candidate feature for the function are provided: Looking for already existing features in the genome using a protein-based BLAST search, and trying to find candidates via a translated BLAST search against the DNA sequence of the whole genome. In both cases, | + | In this page you can search for a function in a genome in the context of a subsystem. Two ways of looking for a candidate feature for the function are provided: Looking for already existing features in the genome using a protein-based BLAST search, and trying to find candidates via a translated BLAST search against the DNA sequence of the whole genome. In both cases, '''reference feature(s)''' are selected (or given to the page) from already existing features with assigned with the functional role that are part of the given subsystem. |
=== Candidate genes found by Similarity (BLAST)? === | === Candidate genes found by Similarity (BLAST)? === | ||
| + | |||
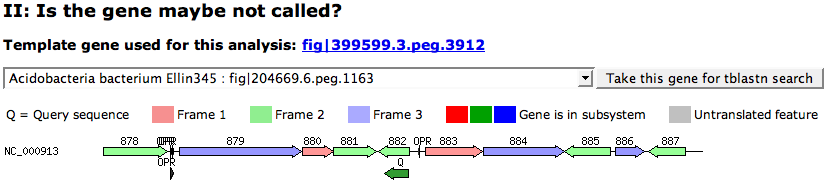
| + | This part looks for [[Glossary#Similarities|Similarities]] of all '''reference feature''' (features in the subsystem that are assigned with the function) to the features of the selected genome. The output [[WebComponent/Table|table]] lists the E-Value ('''P-Sc'''), the reference feature that was hit ('''PEG'''), the length ('''Len''') of the hit feature, the current function of the hit feature ('''Current fn'''), as well as the query feature ('''Matched peg'''), its length ('''Len'''), and its '''Function'''. | ||
[[Image:SearchGene1.png]] | [[Image:SearchGene1.png]] | ||
Revision as of 10:41, 2 December 2008
Search Gene
In this page you can search for a function in a genome in the context of a subsystem. Two ways of looking for a candidate feature for the function are provided: Looking for already existing features in the genome using a protein-based BLAST search, and trying to find candidates via a translated BLAST search against the DNA sequence of the whole genome. In both cases, reference feature(s) are selected (or given to the page) from already existing features with assigned with the functional role that are part of the given subsystem.
Candidate genes found by Similarity (BLAST)?
This part looks for Similarities of all reference feature (features in the subsystem that are assigned with the function) to the features of the selected genome. The output table lists the E-Value (P-Sc), the reference feature that was hit (PEG), the length (Len) of the hit feature, the current function of the hit feature (Current fn), as well as the query feature (Matched peg), its length (Len), and its Function.