Difference between revisions of "SEED Viewer Manual/GenomeBrowser"
| (9 intermediate revisions by 2 users not shown) | |||
| Line 7: | Line 7: | ||
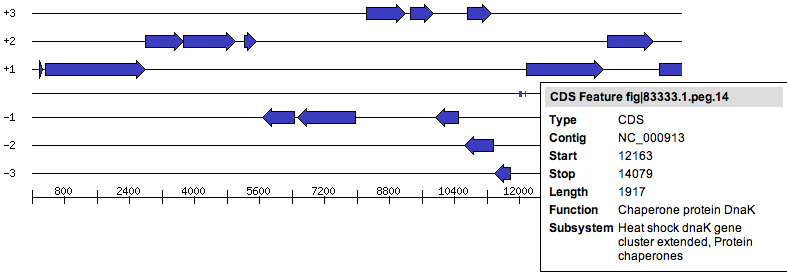
The six frame view represents the six reading frames for proteins (-3, -2, -1, 1, 2, 3). Blue arrows are printed representing protein features. Their direction depicts the strand of the protein (- or +). | The six frame view represents the six reading frames for proteins (-3, -2, -1, 1, 2, 3). Blue arrows are printed representing protein features. Their direction depicts the strand of the protein (- or +). | ||
| − | As RNA features (tRNAs, rRNAs, or other genomic features like binding sites) have no reading frames | + | As RNA features (tRNAs, rRNAs, or other genomic features like binding sites) have no reading frames. They can be found in the middle line of the image, represented by little blue boxes. |
Hovering over a feature will show a tooltip containing information about the feature, including the name, the position in the genome, the [[Glossary#Functional_role|functional role]] and the [[Glossary#Subsystem|subsystem(s)]] it's in. | Hovering over a feature will show a tooltip containing information about the feature, including the name, the position in the genome, the [[Glossary#Functional_role|functional role]] and the [[Glossary#Subsystem|subsystem(s)]] it's in. | ||
| Line 15: | Line 15: | ||
=== The Control TabView === | === The Control TabView === | ||
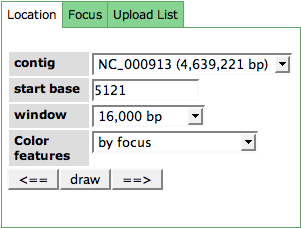
| − | Controlling the Six Frame View can be done using the first tab of the Control TabView. You can choose a location the of the genome you want to view by selecting a contig in the '''contig''' drop down menu and stating a '''start position''' on that contig. The zoom of the window can be changed using the '''window''' drop down box. In addition, features can be colored by different metaphors: by subsystem, by filter options of the Feature Table (the features that are present in the table after filtering it will be colored) or by a user defined list you can specify in the '''Upload List''' tab. | + | Controlling the Six Frame View can be done using the first tab ('''Location''') of the Control TabView. You can choose a location the of the genome you want to view by selecting a contig in the '''contig''' drop down menu and stating a '''start position''' on that contig. The zoom of the window can be changed using the '''window''' drop down box. |
| + | |||
| + | In addition, features can be colored by different metaphors: by subsystem, by filter options of the Feature Table (the features that are present in the table after filtering it will be colored) or by a user defined list you can specify in the '''Upload List''' tab. | ||
| + | |||
| + | Click the button '''Draw''' after making your selections. The arrows '''<=''' and '''=>''' will shift the window over the contig sequence. | ||
| + | |||
| + | If you have selected a feature in the Six Frame View, the second tab ('''Focus''') will show you information about the feature (the same information you can find in the tooltip by hovering over the feature). Additionally, you will get three buttons that lead to pages that show more detailed information about the selected feature. | ||
| + | |||
| + | The button '''zoom to sequence''' will show you the '''[[SEED_Viewer_Manual/ContigView|DNA to Protein]]''' page depicting a detailed sequence view including the DNA sequence, a six frame view of the translation to protein and other features of the sequence in the area of the selected feature. | ||
| + | |||
| + | Clicking '''details page''' leads to the [[SEED_Viewer_Manual/Annotation|Annotation]] page of the feature. You will see all known details about the feature, as well as the [[SEED_Viewer_Manual/Annotation#CompareRegions|Compare regions view]] centered on the feature. | ||
| + | |||
| + | The '''evidence''' button leads to the [[SEED_Viewer_Manual/Evidence|Evidence]] page showing evidence for the annotation of the feature in form of Similarities and protein domains. | ||
| + | |||
| + | '''Upload a list''' - will let you upload a list of locations of BLAST hits of the form (Contig, Start, Stop, ID). The list has to be in plain text format, meaning not an Excel table. If you have an Excel table containing the information, save the list as '''text (tab delimited)''' in Excel. If you have successfully uploaded a list, you will now be able to navigate the regions in your list. They are shown on the middle line in the Six Frame View in form of little boxes. | ||
[[Image:GenomeBrowserLoc.png]] | [[Image:GenomeBrowserLoc.png]] | ||
| + | |||
| + | === The Feature Table === | ||
| + | |||
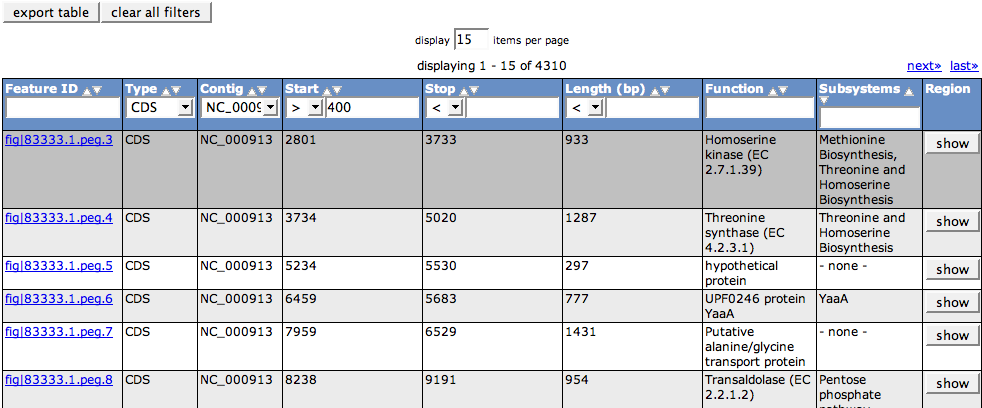
| + | The Feature Table shows all features present in your organism in a [[WebComponents/Table|table]]. Information you can see for a feature are its ID, Type (e.g. CDS, RNA and others), its locations (Contig, Start, Stop) and length as well as the functional role it's annotated with and the subsystems it belongs to. The button in the last column (Region) will center the Six Frame View on this feature and select it. | ||
[[Image:GenomeBrowserFeat.png]] | [[Image:GenomeBrowserFeat.png]] | ||
Latest revision as of 05:58, 5 December 2008
Genome Browser
The Genome Browser enables you to view the features of a genome in their genomic context. The page is divided into three parts, a Control TabView, the Six Frame View, and a table showing all features of an organism.
The Six Frame View
The six frame view represents the six reading frames for proteins (-3, -2, -1, 1, 2, 3). Blue arrows are printed representing protein features. Their direction depicts the strand of the protein (- or +).
As RNA features (tRNAs, rRNAs, or other genomic features like binding sites) have no reading frames. They can be found in the middle line of the image, represented by little blue boxes.
Hovering over a feature will show a tooltip containing information about the feature, including the name, the position in the genome, the functional role and the subsystem(s) it's in.
The Control TabView
Controlling the Six Frame View can be done using the first tab (Location) of the Control TabView. You can choose a location the of the genome you want to view by selecting a contig in the contig drop down menu and stating a start position on that contig. The zoom of the window can be changed using the window drop down box.
In addition, features can be colored by different metaphors: by subsystem, by filter options of the Feature Table (the features that are present in the table after filtering it will be colored) or by a user defined list you can specify in the Upload List tab.
Click the button Draw after making your selections. The arrows <= and => will shift the window over the contig sequence.
If you have selected a feature in the Six Frame View, the second tab (Focus) will show you information about the feature (the same information you can find in the tooltip by hovering over the feature). Additionally, you will get three buttons that lead to pages that show more detailed information about the selected feature.
The button zoom to sequence will show you the DNA to Protein page depicting a detailed sequence view including the DNA sequence, a six frame view of the translation to protein and other features of the sequence in the area of the selected feature.
Clicking details page leads to the Annotation page of the feature. You will see all known details about the feature, as well as the Compare regions view centered on the feature.
The evidence button leads to the Evidence page showing evidence for the annotation of the feature in form of Similarities and protein domains.
Upload a list - will let you upload a list of locations of BLAST hits of the form (Contig, Start, Stop, ID). The list has to be in plain text format, meaning not an Excel table. If you have an Excel table containing the information, save the list as text (tab delimited) in Excel. If you have successfully uploaded a list, you will now be able to navigate the regions in your list. They are shown on the middle line in the Six Frame View in form of little boxes.
The Feature Table
The Feature Table shows all features present in your organism in a table. Information you can see for a feature are its ID, Type (e.g. CDS, RNA and others), its locations (Contig, Start, Stop) and length as well as the functional role it's annotated with and the subsystems it belongs to. The button in the last column (Region) will center the Six Frame View on this feature and select it.