Difference between revisions of "SEED Viewer Manual/FIGfamViewer"
| (4 intermediate revisions by the same user not shown) | |||
| Line 3: | Line 3: | ||
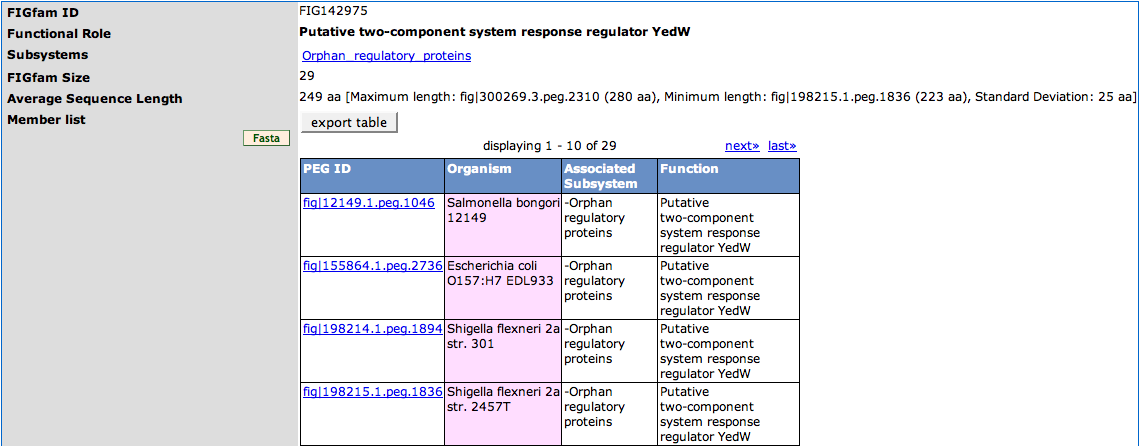
The table on the top of the page displays the '''FIGfam ID''' and the '''Functional Role''' associated to the FIGfam. Subsystems that are associated with members of the FIGfam are displayed in the third row ('''Subsystems''') of the table. They are linked to the [[SEED_Viewer_Manual/Subsystems|Subsystems Page]] of the subsystem. The '''FIGfam Size''' is the number of members of the FIGfam. The '''Average Sequence Length''' is the medium of aminoacids sequence length of the members of the FIGfam. Behind this there is a short statistics telling you the member with the '''Maximum length''', '''Minimum length''' and the '''Standard Deviation'''. | The table on the top of the page displays the '''FIGfam ID''' and the '''Functional Role''' associated to the FIGfam. Subsystems that are associated with members of the FIGfam are displayed in the third row ('''Subsystems''') of the table. They are linked to the [[SEED_Viewer_Manual/Subsystems|Subsystems Page]] of the subsystem. The '''FIGfam Size''' is the number of members of the FIGfam. The '''Average Sequence Length''' is the medium of aminoacids sequence length of the members of the FIGfam. Behind this there is a short statistics telling you the member with the '''Maximum length''', '''Minimum length''' and the '''Standard Deviation'''. | ||
| − | The '''Member list''' is a [[WebComponents/Table|table]] showing the '''ID''' of the member, the '''Organism''' it belongs to, '''Associated Subsystem''' (there can be more than one) and the function of the member. | + | The '''Member list''' is a [[WebComponents/Table|table]] showing the '''ID''' of the member (the link leads to the [[SEED_Viewer_Manual/Annotation|Annotation Page]]), the '''Organism''' it belongs to, '''Associated Subsystem''' (there can be more than one) and the function of the member. You can export the list in ''tab-separated format'' using the '''export table''' button. For downloading a fasta file of the protein sequences of the members, press the '''Fasta''' button on the left. |
[[Image:FIGfamViewerTable.png]] | [[Image:FIGfamViewerTable.png]] | ||
| + | |||
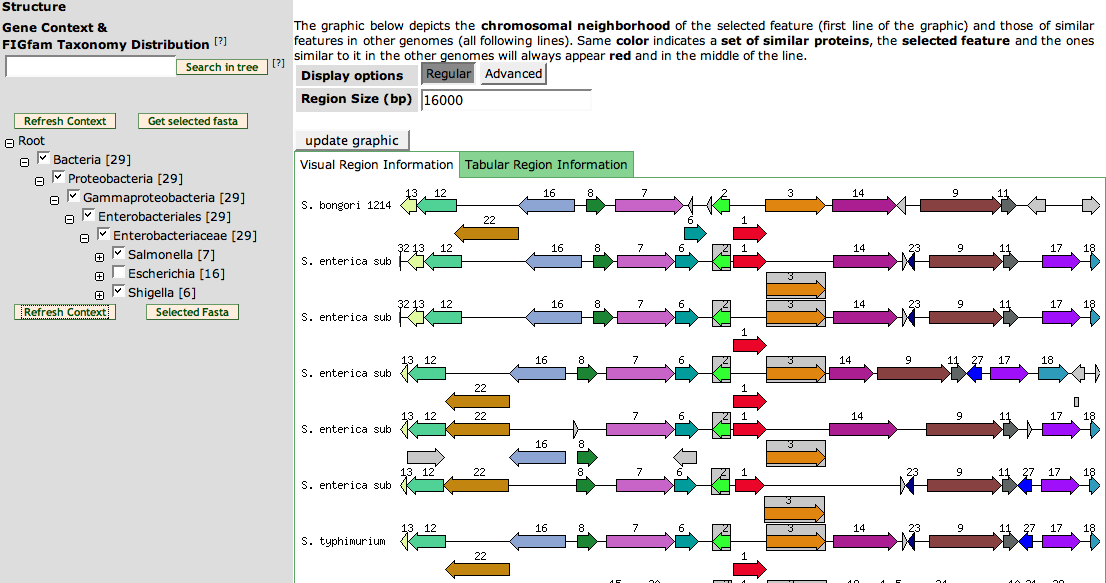
| + | The bottom part of the page shows a '''Compare Regions View''' of the members of the FIGfam. The right graphics is a '''Compare Regions''' just like the one on the [[SEED_Viewer_Manual/Annotation|Annotation Page]]. Click [[SEED_Viewer_Manual/Annotation#Compare Regions|here]] to get a learn more. | ||
| + | |||
| + | The left part is used to filter the Compare Regions View. It is an phylogenetic tree showing only the phyla with members for the FIGfam. Click the '''+''' signs to expand and '''-''' signs to collapse a part. You can deselect check boxes to remove the organisms from the Compare Regions View. Press '''Refresh Context''' to update the View. | ||
| + | |||
| + | If you want to find a member of the FIGfam, you can put the ID into the text field on top of the tree. Clicking the '''search in tree''' button will expand the tree up to the position of the FIGfam member. | ||
| + | |||
| + | The button '''get selected fasta''' will let you download the fasta sequences of all members in the organisms you have selected in the tree. | ||
| + | |||
| + | [[Image:FIGfamViewerTree.png]] | ||
Latest revision as of 15:07, 3 December 2008
FIGfam Viewer
The table on the top of the page displays the FIGfam ID and the Functional Role associated to the FIGfam. Subsystems that are associated with members of the FIGfam are displayed in the third row (Subsystems) of the table. They are linked to the Subsystems Page of the subsystem. The FIGfam Size is the number of members of the FIGfam. The Average Sequence Length is the medium of aminoacids sequence length of the members of the FIGfam. Behind this there is a short statistics telling you the member with the Maximum length, Minimum length and the Standard Deviation.
The Member list is a table showing the ID of the member (the link leads to the Annotation Page), the Organism it belongs to, Associated Subsystem (there can be more than one) and the function of the member. You can export the list in tab-separated format using the export table button. For downloading a fasta file of the protein sequences of the members, press the Fasta button on the left.
The bottom part of the page shows a Compare Regions View of the members of the FIGfam. The right graphics is a Compare Regions just like the one on the Annotation Page. Click here to get a learn more.
The left part is used to filter the Compare Regions View. It is an phylogenetic tree showing only the phyla with members for the FIGfam. Click the + signs to expand and - signs to collapse a part. You can deselect check boxes to remove the organisms from the Compare Regions View. Press Refresh Context to update the View.
If you want to find a member of the FIGfam, you can put the ID into the text field on top of the tree. Clicking the search in tree button will expand the tree up to the position of the FIGfam member.
The button get selected fasta will let you download the fasta sequences of all members in the organisms you have selected in the tree.