Difference between revisions of "SEED Viewer Manual/Editing Capabilities/ChromosomalClusters"
| Line 3: | Line 3: | ||
This page can be accessed from a [[SEED_Viewer_Manual/Annotation#Chromosomal Clusters|Chromosomal Clusters]] view, using the button '''annotate clusters''' (only visible if you have editing rights for the genome). | This page can be accessed from a [[SEED_Viewer_Manual/Annotation#Chromosomal Clusters|Chromosomal Clusters]] view, using the button '''annotate clusters''' (only visible if you have editing rights for the genome). | ||
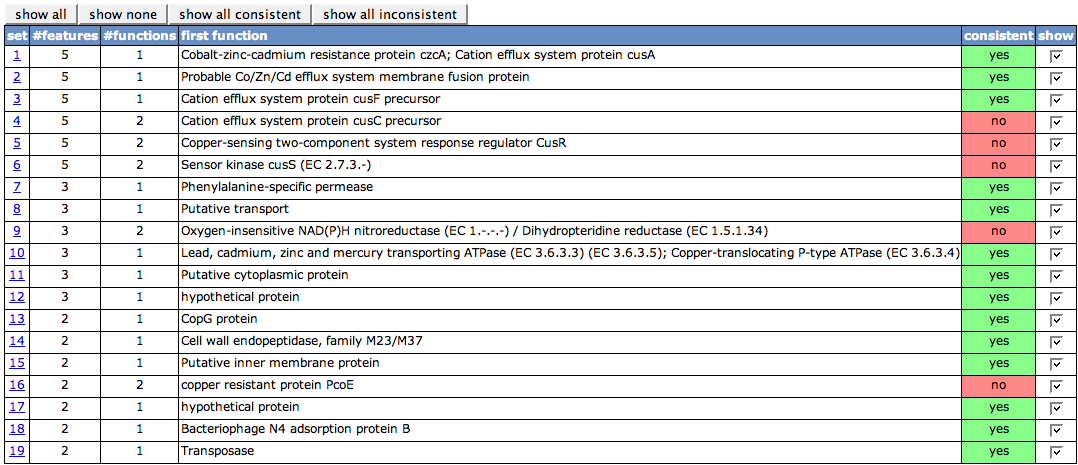
| − | The first table on the page lists all sets of protein visible in the Chromosomal Clusters view. The set numbers in the first column are the set numbers from the view. The second column ('''#features''') shows the number of features in the set. '''#functions''' depicts how many different functions the features in the set have. Whenever there is more than one function, the cell in the next column will be painted in red, as it is not '''consistent''', else green. With the check boxes in the last column, you can turn on and off the details tables for each of the sets. | + | The first table on the page lists all sets of protein visible in the Chromosomal Clusters view. The set numbers in the first column are the set numbers from the view. The second column ('''#features''') shows the number of features in the set. '''#functions''' depicts how many different functions the features in the set have. One of the functions is displayed in the column '''first function'''. Whenever there is more than one function, the cell in the next column will be painted in red, as it is not '''consistent''', else green. With the check boxes in the last column, you can turn on and off the details tables for each of the sets. |
[[Image:Chromosomal1.png]] | [[Image:Chromosomal1.png]] | ||
As a default, for each set you will get a details table in the following. These list all the features in the set. The set number is displayed in the first column. '''Organism''' is the genome the feature stems from. If there are more than one occurrence of features in the set for one genome, they are numbered in the '''Occ''' column. If there is a [http://www.uniprot.org UniProt] alias for the feature, the ID and the function of this alias will be shown in the next two columns. | As a default, for each set you will get a details table in the following. These list all the features in the set. The set number is displayed in the first column. '''Organism''' is the genome the feature stems from. If there are more than one occurrence of features in the set for one genome, they are numbered in the '''Occ''' column. If there is a [http://www.uniprot.org UniProt] alias for the feature, the ID and the function of this alias will be shown in the next two columns. | ||
Revision as of 05:48, 8 December 2008
Editing Capabilities - Chromosomal Clusters
This page can be accessed from a Chromosomal Clusters view, using the button annotate clusters (only visible if you have editing rights for the genome).
The first table on the page lists all sets of protein visible in the Chromosomal Clusters view. The set numbers in the first column are the set numbers from the view. The second column (#features) shows the number of features in the set. #functions depicts how many different functions the features in the set have. One of the functions is displayed in the column first function. Whenever there is more than one function, the cell in the next column will be painted in red, as it is not consistent, else green. With the check boxes in the last column, you can turn on and off the details tables for each of the sets.
As a default, for each set you will get a details table in the following. These list all the features in the set. The set number is displayed in the first column. Organism is the genome the feature stems from. If there are more than one occurrence of features in the set for one genome, they are numbered in the Occ column. If there is a UniProt alias for the feature, the ID and the function of this alias will be shown in the next two columns.