Difference between revisions of "Home of the SEED"
FolkerMeyer (talk | contribs) m |
m |
||
| (34 intermediate revisions by 7 users not shown) | |||
| Line 1: | Line 1: | ||
| − | With the growing number of genomes | + | With the growing number of available genomes, the need for an environment to support effective comparative analysis increases. The original SEED Project was started in 2003 by the [http://thefig.info Fellowship for Interpretation of Genomes (FIG)] as a largely unfunded open source effort. Argonne National Laboratory and the University of Chicago joined the project, and now much of the activity occurs at those two institutions (as well as the University of Illinois at Urbana-Champaign, Hope college, San Diego State University, the Burnham Institute and a number of other institutions). The cooperative effort focuses on the development of the comparative genomics environment called the SEED and, more importantly, on the development of curated genomic data. |
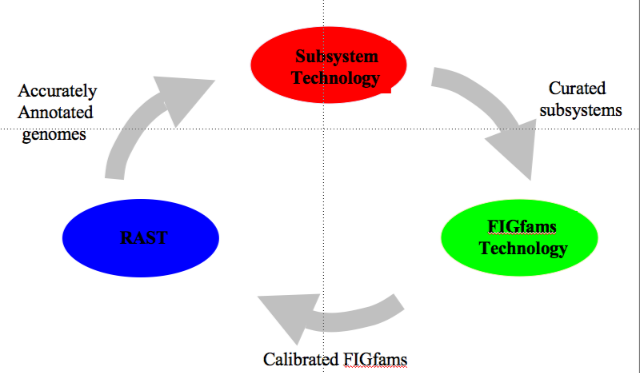
| − | + | [[Image:Data_lifecycle3.png|frame|thumbnail|50px|Flow of data in the SEED.]] | |
| − | + | Curation of genomic data ([[Glossary#Annotation|annotation]]) is done via the curation of [[Glossary#Subsystem|subsystems]] by an expert annotator across many genomes, not on a gene by gene basis. This is also detailed in our [[Annotating_1000_genomes|manifesto]]. From the curated subsystems we extract a set of freely available protein families ([http://seed-viewer.theseed.org/seedviewer.cgi?page=FigFamViewer FIGfams]). These FIGfams form the core component of our RAST automated annotation technology. | |
| + | Answering numerous requests for automatic Seed-Quality annotations for more or less complete bacterial and archaeal genomes, we have established the free [http://rast.nmpdr.org RAST-Server] (RAST=Rapid Annotation using Subsytems Technology). Using similar technology, we make the [http://metagenomics.nmpdr.org Metagenomics-RAST-Server] freely available. We also provide a [http://seed-viewer.theseed.org/ SEED-Viewer] that allows read-only access to the latest curated data sets. For users interested in editing and learning how to use the system, we provide the [http://theseed.uchicago.edu/FIG/index.cgi Trial-SEED]. | ||
| + | We make all our software and data available for download via [ftp://ftp.theseed.org], also see our [[DownloadPage]] page. | ||
| − | + | * We request that groups using the SEED cite: Overbeek et al., [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=pubmed&dopt=Abstract&list_uids=16214803&query_hl=2&itool=pubmed_docsum|Nucleic Nucleic Acids Res 33(17)], 2005 ([http://www.theseed.org/SubsystemPaperSupplementalMaterial/index.html Supplementary material]) | |
| − | * We | + | * We request that groups using the RAST server cite: Aziz et al., [http://www.ncbi.nlm.nih.gov/pubmed/18261238?ordinalpos=1&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_RVDocSum| BMC Genomics], 2008 ([http://www.theseed.org/RASTPaperSupplementalMaterial/index.html Supplementary material]) |
| − | + | * Our approaches to annotation, gene calling, etc. are outlined in a series of [http://www.nmpdr.org/FIG/wiki/view.cgi/SOP/WebHome| Standard Operating Procedures]. | |
| − | * Our approaches to annotation, gene calling etc are outlined in a series of [ | + | * Read about how other groups have used our tools in their research on our [http://www.nmpdr.org/FIG/wiki/view.cgi/Main/Publications#Using Publications] page. |
Latest revision as of 15:52, 11 June 2010
With the growing number of available genomes, the need for an environment to support effective comparative analysis increases. The original SEED Project was started in 2003 by the Fellowship for Interpretation of Genomes (FIG) as a largely unfunded open source effort. Argonne National Laboratory and the University of Chicago joined the project, and now much of the activity occurs at those two institutions (as well as the University of Illinois at Urbana-Champaign, Hope college, San Diego State University, the Burnham Institute and a number of other institutions). The cooperative effort focuses on the development of the comparative genomics environment called the SEED and, more importantly, on the development of curated genomic data.
Curation of genomic data (annotation) is done via the curation of subsystems by an expert annotator across many genomes, not on a gene by gene basis. This is also detailed in our manifesto. From the curated subsystems we extract a set of freely available protein families (FIGfams). These FIGfams form the core component of our RAST automated annotation technology.
Answering numerous requests for automatic Seed-Quality annotations for more or less complete bacterial and archaeal genomes, we have established the free RAST-Server (RAST=Rapid Annotation using Subsytems Technology). Using similar technology, we make the Metagenomics-RAST-Server freely available. We also provide a SEED-Viewer that allows read-only access to the latest curated data sets. For users interested in editing and learning how to use the system, we provide the Trial-SEED.
We make all our software and data available for download via [1], also see our DownloadPage page.
- We request that groups using the SEED cite: Overbeek et al., Nucleic Acids Res 33(17), 2005 (Supplementary material)
- We request that groups using the RAST server cite: Aziz et al., BMC Genomics, 2008 (Supplementary material)
- Our approaches to annotation, gene calling, etc. are outlined in a series of Standard Operating Procedures.
- Read about how other groups have used our tools in their research on our Publications page.